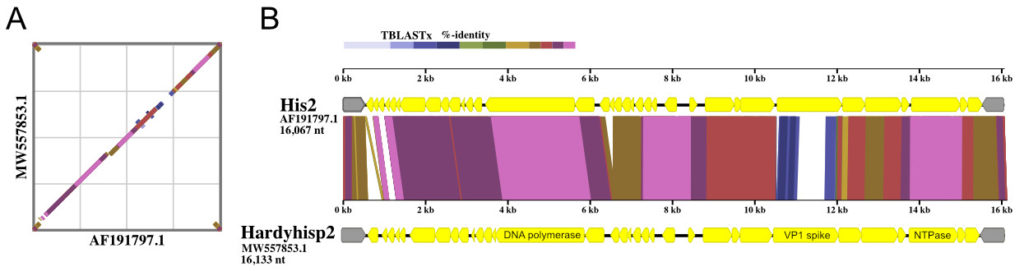
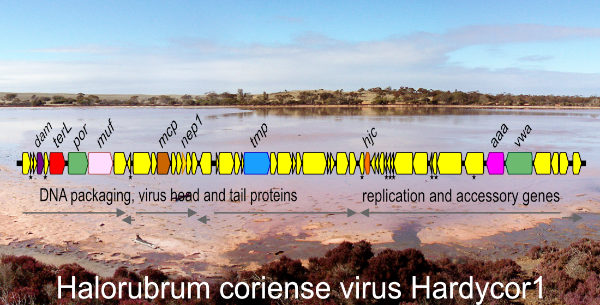
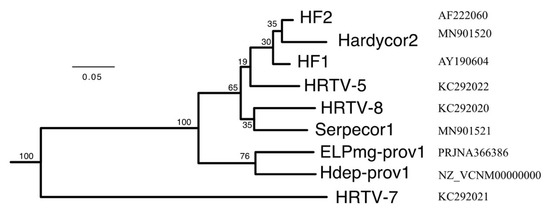
In two earlier studies of Haloquadratum walsbyi [1,2], Australian isolates were often found to harbor small cryptic plasmids of the pL6-family, approximately 6 kb in size (see diagram above). Only five examples of these plasmids had been previously described but they are peculiar because they show no homology to other haloarchaeal plasmids yet display a highly conserved gene arrangement. Intriguingly, they encode a replicase very similar to those found in betapleolipoviruses. In the current study [3], fifteen additional plasmids were reconstructed by mining the metagenomes of hypersaline sites in four countries: Argentina, Australia, Puerto Rico, and Spain. Overall, the average plasmid size was 6002 bp, and the gene arrangement remained well conserved. Many metagenomic CRISPR spacers were found to match sequences in the fifteen reconstructed plasmids, indicating frequent invasion of haloarchaea. A surprise finding was that strand-specific metatranscriptome (RNA-seq) data from other published studies could be used to map transcripts produced by two plasmids, confirming the active transcription of coding sequences and also high levels of counter-transcripts. It remains unclear whether these fascinating, highly conserved plasmids are viruses or perhaps virus satellites, but they are widespread and probably extend beyond Haloquadratum.